About
The PLVis Repository
The PLVis Repository is a page dedicated to showcasing the similarities and differences between the proteomes of different species along the Tree of Life using Protein Language Model (PLM) embeddings.
In our previous manuscript titled “The Protein Language Visualizer: Sequence Similarity Networks for the Era of Language Models”, we demonstrated how 2-dimensional reduced embeddings can be clustered together to find groups of proteins with similar functional and structural features.
We have therefore created this repository as a way to explore the function-structure space of proteins belonging to closely related organisms in the Tree of Life.
For this website, reference proteomes were retrieved from UniProt for the available species in all domains of life.
The proteomes were then organized according to their taxonomic family and visualized together.
Each visualization is accompanied by its corresponding enrichment report, which contains the k-clusters in the comparison that share a specific trait (e.g. Gene, Organism, InterPro ID).
Proteome Comparison Pipeline
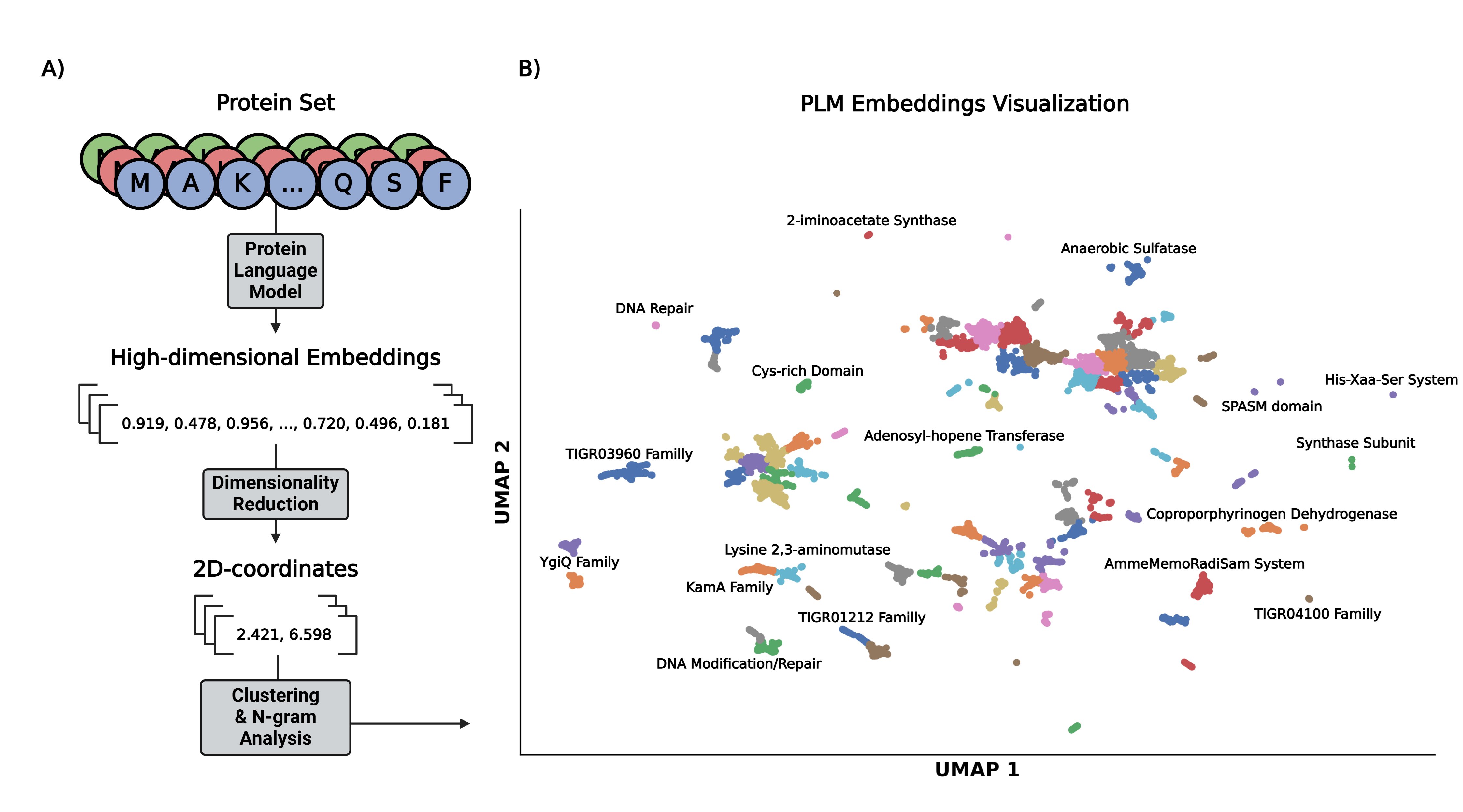
All proteome comparisons featured in the PLVis repository are based on the pipeline featured in the aforementioned manuscript.
First, the high-dimensional embeddings for each of the sequences in the set are calculated using a PLM (e.g. ESM2, ProtT5).
Next, a dimensionality reduction algorithm (e.g. UMAP, tSNE, TriMAP) decreases the number of dimensions for each embedding to two.
After that, a clustering method (e.g. k-Means, DBSCAN) is used to identify groupings of proteins in the 2-dimensional visualization.
Lastly, an n-gram analysis is performed to generate a name for each cluster based on the two most repeated words found in the corresponding protein names.
The proteome comparisons found in the PLVis Repository were generated using ProtT5 embeddings, UMAP reduction and k-Means clustering.
You can find the corresponding manuscript as well as a Google Colab notebook to generate a PLVis projection using any protein set in the following links:
Manuscript:
https://www.biorxiv.org/content/10.1101/2024.11.19.624229v2
PLVis Colab:
https://colab.research.google.com/drive/1s5ug8CYaJ4unJIElxfLzcsvxUWPNqWfD?usp=sharing
Curated Comparisons
The 'Curated Comparisons' section comprises different case studies from guest undergraduate students at the Jinich Lab during the ENLACE program. Unlike the other comparisons in the repository, these visualizations are not focused on specific families in the Tree of Life, instead, they compare organisms with pathological relationships or shared functions. All proteome comparisons in this section feature a written report of the visualization, highlighting sections (k-clusters) of interest for the specific study.